Glutamate Dehydrogenase: an enzyme at the heart of energy metabolism.
 Introduction. The enzyme glutamate dehydrogenase (pictured RHS) is ubiquitous in eukaryotes; forming a link between energy production via the Krebs (or Tricarboxylic Acid [TCA] Cycle. Early investigations described the substrates and products, but it was between 1960 and 1990 that most of the molecular work on this enzyme was published. The pioneering enzymological work was carried out on the enzyme from bovine liver (beef liver) and in particular the laboratories of Carl Frieden (now at the University of Washington in St. Louis) and Paul Engel (now at University College Dublin); more recently a great deal of crystallography has been carried out on bacterial GDHs in the laboratories of David Rice and Pat Baker at the University of Sheffield. The first GDH to be crystallised was isolated from an anaerobic bacterium, Clostridium symbiosum at Sheffield in the late 1980s by Engel, Rice and Hornby. This work provided a framework for understanding the catalytic mechanism of this enzyme and has been followed more recently by the determination of structures for vertebrate GDHs, in 2002.
Introduction. The enzyme glutamate dehydrogenase (pictured RHS) is ubiquitous in eukaryotes; forming a link between energy production via the Krebs (or Tricarboxylic Acid [TCA] Cycle. Early investigations described the substrates and products, but it was between 1960 and 1990 that most of the molecular work on this enzyme was published. The pioneering enzymological work was carried out on the enzyme from bovine liver (beef liver) and in particular the laboratories of Carl Frieden (now at the University of Washington in St. Louis) and Paul Engel (now at University College Dublin); more recently a great deal of crystallography has been carried out on bacterial GDHs in the laboratories of David Rice and Pat Baker at the University of Sheffield. The first GDH to be crystallised was isolated from an anaerobic bacterium, Clostridium symbiosum at Sheffield in the late 1980s by Engel, Rice and Hornby. This work provided a framework for understanding the catalytic mechanism of this enzyme and has been followed more recently by the determination of structures for vertebrate GDHs, in 2002.
The reaction catalysed by GDH (sometimes abbreviated to GluDH or GLDH), lies in the direction of ammonia assimilation. However, the equilibrium constant makes it possible to measure both directions, and in this project we shall mainly utilise the oxidative deamination of glutamate for our measurements.
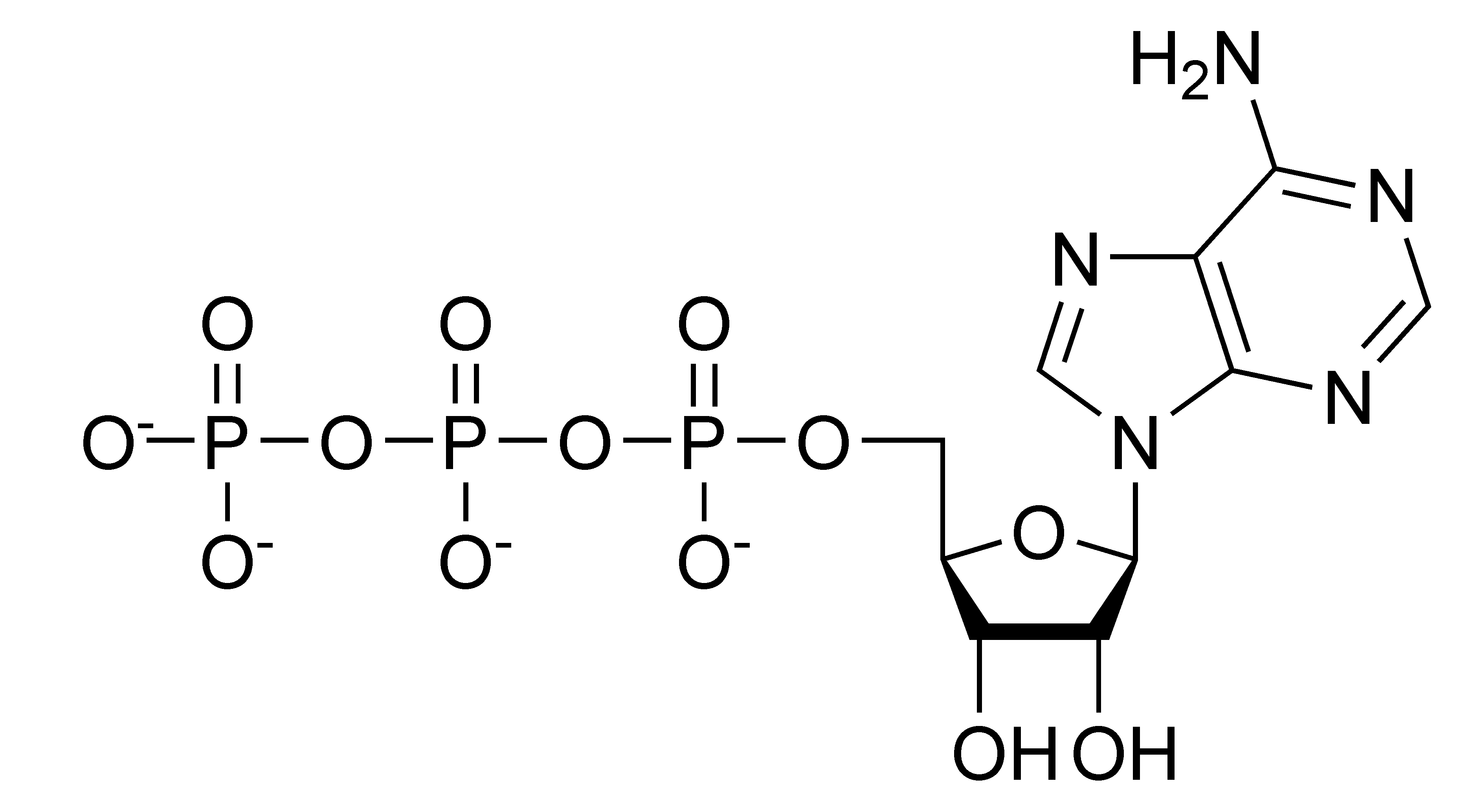
Glutamate +NAD(P)+ ↔ 2-oxoglutarate + NH4+ + NADH + H+
Our understanding of the conversion of foods such as carbohydrates (eg glucose), lipids (fats) and proteins was one of the major achievements of 20th century Biochemistry and Physiology. Later developments in our understanding of the concepts of “building blocks” and the biosynthesis of our own body fats, carbohydrates and proteins helped us to appreciate the complex systems that cells, tissues and organisms employ to not only produce energy, but also to grow and develop. The recent term “Systems Biology” is used by many to describe the integrated sets of reactions and pathways that define the function of an organism at a range of hierarchical levels.
 For example, you might be interested in ATP synthesis. This is the product of oxidative phosphorylation in the mitochondria of vertebrates. However, ATP is also produced as a product of a large number of enzyme catalysed reactions. Moreover, ATP is utilised in the biosynthesis of glycogen, the synthesis of nucleic acids and is a regulator of many biochemical activities (the transfer of the terminal phosphate of ATP to certain proteins can selectively turn on (or off) their catalytic function. This suggests that studying individual enzyme catalysed reactions, while important, has to be seen in a wider context. This is probably the most important aspect of “Systems Biology”. GDH catalyses the mobilisation of 2-oxoglutarate (also called α-ketoglutarate) either to generate the amino acid L-glutamate for incorporation into newly synthesised proteins, or to provide a valuable metabolite for neuronal signalling. In the reverse reaction, the enzyme “tops up” the TCA cycle (see top RHS), since the reaction catalysed by 2-oxoglutarate dehydrogenase precedes the formation of succinyl CoA.
For example, you might be interested in ATP synthesis. This is the product of oxidative phosphorylation in the mitochondria of vertebrates. However, ATP is also produced as a product of a large number of enzyme catalysed reactions. Moreover, ATP is utilised in the biosynthesis of glycogen, the synthesis of nucleic acids and is a regulator of many biochemical activities (the transfer of the terminal phosphate of ATP to certain proteins can selectively turn on (or off) their catalytic function. This suggests that studying individual enzyme catalysed reactions, while important, has to be seen in a wider context. This is probably the most important aspect of “Systems Biology”. GDH catalyses the mobilisation of 2-oxoglutarate (also called α-ketoglutarate) either to generate the amino acid L-glutamate for incorporation into newly synthesised proteins, or to provide a valuable metabolite for neuronal signalling. In the reverse reaction, the enzyme “tops up” the TCA cycle (see top RHS), since the reaction catalysed by 2-oxoglutarate dehydrogenase precedes the formation of succinyl CoA.The key information for today's lab class is below. Further details will be discussed during the session: this is just to give you an idea of planning your experiments. You must obtain all of the necessary reagents and equipment as a group from the usual locations in Innovation Lab 1.
Practical enzymology with GDH. In order to fully appreciate how enzyme's catalyse reactions, we need to observe them in action. We are going to begin with preparing the reagents for measuring the conversion of NAD to NADH via the oxidative deamination of glutamate, catalysed by Clostridium symbiosum GDH. The measurements of rates will be discussed in the next blog in some detail, but first I want to discuss the preparations in advance of rate measurements.
The substrates. The substrates are provided as solid sodium salts of glutamate and nicotinamide adenine dinucleotide (NAD+). Both were obtained from Thermo Fisher. You should make notes on the details provided by the manufacturer, including any chemical characteristics, molecular weight and notes on storage requirements.
Each group should prepare 100ml of 200mM glutamate in distilled water. You should measure the pH and aliquot the solution into 10x10ml Falcon tubes for storage.
Similarly, you should prepare 10ml of 10mM NAD+ and store 10x1ml Eppendorfs. No need to measure the pH.
Make sure your samples are clearly labelled and that you have stored the solutions correctly.
The buffer. Prepare a 5x concentrate of PBS and check its final pH. (we shall use lower and higher pH buffers later for measuring the influence of pH on reaction rates.
Standard reaction mixture:
50mM glutamate
1mMNAD+
PBS
Final volume 1ml
Enzyme volume 5ul
Room temperature
The enzyme. You are provided with a suspension of GDH in saturated ammonium sulphate at a concentration of 20mg/ml. The activity of the enzyme preparation must be established by making a series of dilutions. We will use PBS for diluting the enzyme samples, we will then establish the appropriate enzyme dilution for measuring rates and end points reproducibly.
The aim of this week's lab work is to "see enzymes in action"; having demonstrated this, in a reproducible manner, we can begin to investigate how temperature, pH, inhibitors such as heavy metals (which tend to be non-specific) and competitive inhibitors (similar to drugs), influence reaction rates in vitro. These experiments will reinforce the concepts that we have discussed in the seminars. Additional information is available in Unit 13 at the Google Classroom.

.jpg)